Note
Go to the end to download the full example code.
Multiple-kernel ridge refining¶
This example demonstrates how to solve multiple-kernel ridge regression with hyperparameter random search, then refine the results with hyperparameter gradient descent.
import numpy as np
from himalaya.backend import set_backend
from himalaya.kernel_ridge import MultipleKernelRidgeCV
from himalaya.kernel_ridge import Kernelizer
from himalaya.kernel_ridge import ColumnKernelizer
from himalaya.utils import generate_multikernel_dataset
from sklearn.pipeline import make_pipeline
from sklearn import set_config
set_config(display='diagram')
In this example, we use the cupy backend (GPU).
backend = set_backend("cupy", on_error="warn")
/home/runner/work/himalaya/himalaya/himalaya/backend/_utils.py:55: UserWarning: Setting backend to cupy failed: Cupy not installed..Falling back to numpy backend.
warnings.warn(f"Setting backend to {backend} failed: {str(error)}."
Generate a random dataset¶
X_train : array of shape (n_samples_train, n_features)
X_test : array of shape (n_samples_test, n_features)
Y_train : array of shape (n_samples_train, n_targets)
Y_test : array of shape (n_samples_test, n_targets)
(X_train, X_test, Y_train, Y_test, kernel_weights,
n_features_list) = generate_multikernel_dataset(n_kernels=4, n_targets=50,
n_samples_train=600,
n_samples_test=300,
random_state=42)
feature_names = [f"Feature space {ii}" for ii in range(len(n_features_list))]
Prepare the pipeline¶
# Find the start and end of each feature space X in Xs
start_and_end = np.concatenate([[0], np.cumsum(n_features_list)])
slices = [
slice(start, end)
for start, end in zip(start_and_end[:-1], start_and_end[1:])
]
# Create a different ``Kernelizer`` for each feature space.
kernelizers = [("space %d" % ii, Kernelizer(), slice_)
for ii, slice_ in enumerate(slices)]
column_kernelizer = ColumnKernelizer(kernelizers)
Define the random-search model¶
We use very few iteration on purpose, to make the random search suboptimal, and refine it with hyperparameter gradient descent.
solver_params = dict(n_iter=5, alphas=np.logspace(-10, 10, 41))
model_1 = MultipleKernelRidgeCV(kernels="precomputed", solver="random_search",
solver_params=solver_params, random_state=42)
pipe_1 = make_pipeline(column_kernelizer, model_1)
# Fit the model on all targets
pipe_1.fit(X_train, Y_train)
[ ] 0% | 0.00 sec | 5 random sampling with cv |
[...... ] 20% | 1.32 sec | 5 random sampling with cv | 0.75 it/s, ETA: 00:00:05
[............ ] 40% | 2.85 sec | 5 random sampling with cv | 0.70 it/s, ETA: 00:00:04
[.................. ] 60% | 4.27 sec | 5 random sampling with cv | 0.70 it/s, ETA: 00:00:02
[........................ ] 80% | 5.69 sec | 5 random sampling with cv | 0.70 it/s, ETA: 00:00:01
[..............................] 100% | 7.07 sec | 5 random sampling with cv | 0.71 it/s, ETA: 00:00:00
Define the gradient-descent model¶
solver_params = dict(max_iter=10, hyper_gradient_method="direct",
max_iter_inner_hyper=10,
initial_deltas="here_will_go_the_previous_deltas")
model_2 = MultipleKernelRidgeCV(kernels="precomputed", solver="hyper_gradient",
solver_params=solver_params)
pipe_2 = make_pipeline(column_kernelizer, model_2)
Use the random-search to initialize the gradient-descent¶
# We might want to refine only the best predicting targets, since the
# hyperparameter gradient descent is less efficient over many targets.
top = 60 # top 60%
best_cv_scores = backend.to_numpy(pipe_1[-1].cv_scores_.max(0))
mask = best_cv_scores > np.percentile(best_cv_scores, 100 - top)
pipe_2[-1].solver_params['initial_deltas'] = pipe_1[-1].deltas_[:, mask]
pipe_2.fit(X_train, Y_train[:, mask])
[ ] 0% | 0.00 sec | hypergradient_direct |
[ ] 0% | 0.00 sec | hypergradient_direct |
[... ] 10% | 3.20 sec | hypergradient_direct | 0.31 it/s, ETA: 00:00:28
[...... ] 20% | 3.92 sec | hypergradient_direct | 0.51 it/s, ETA: 00:00:15
[......... ] 30% | 4.67 sec | hypergradient_direct | 0.64 it/s, ETA: 00:00:10
[............ ] 40% | 5.41 sec | hypergradient_direct | 0.74 it/s, ETA: 00:00:08
[............... ] 50% | 6.15 sec | hypergradient_direct | 0.81 it/s, ETA: 00:00:06
[.................. ] 60% | 6.89 sec | hypergradient_direct | 0.87 it/s, ETA: 00:00:04
[..................... ] 70% | 7.63 sec | hypergradient_direct | 0.92 it/s, ETA: 00:00:03
[........................ ] 80% | 8.37 sec | hypergradient_direct | 0.96 it/s, ETA: 00:00:02
[........................... ] 90% | 9.10 sec | hypergradient_direct | 0.99 it/s, ETA: 00:00:01
[..............................] 100% | 9.89 sec | hypergradient_direct | 1.01 it/s, ETA: 00:00:00
Compute predictions on a test set¶
import matplotlib.pyplot as plt
# use the first model for all targets
test_scores_1 = pipe_1.score(X_test, Y_test)
# use the second model for the refined targets
test_scores_2 = backend.copy(test_scores_1)
test_scores_2[mask] = pipe_2.score(X_test, Y_test[:, mask])
test_scores_1 = backend.to_numpy(test_scores_1)
test_scores_2 = backend.to_numpy(test_scores_2)
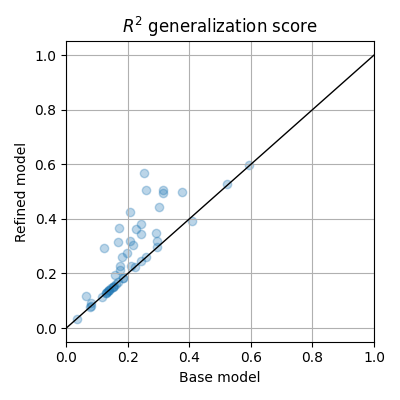
plt.figure(figsize=(4, 4))
plt.scatter(test_scores_1, test_scores_2, alpha=0.3)
plt.xlim(0, 1)
plt.plot(plt.xlim(), plt.xlim(), color='k', lw=1)
plt.xlabel(r"Base model")
plt.ylabel(r"Refined model")
plt.title("$R^2$ generalization score")
plt.grid()
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 20.918 seconds)